MONAILabel Server Installation
To install MONAI Label along with additional models for higher-level morphological structure segmentation, follow below steps.
Using Docker
Docker prerequisites
To deploy server using Docker you need to install Docker on your computer. This step is different for different operating systems. For this purpose follow the instructions in official Docker Documentation.
Visit the Docker Documentation page and select the OS you have installed.
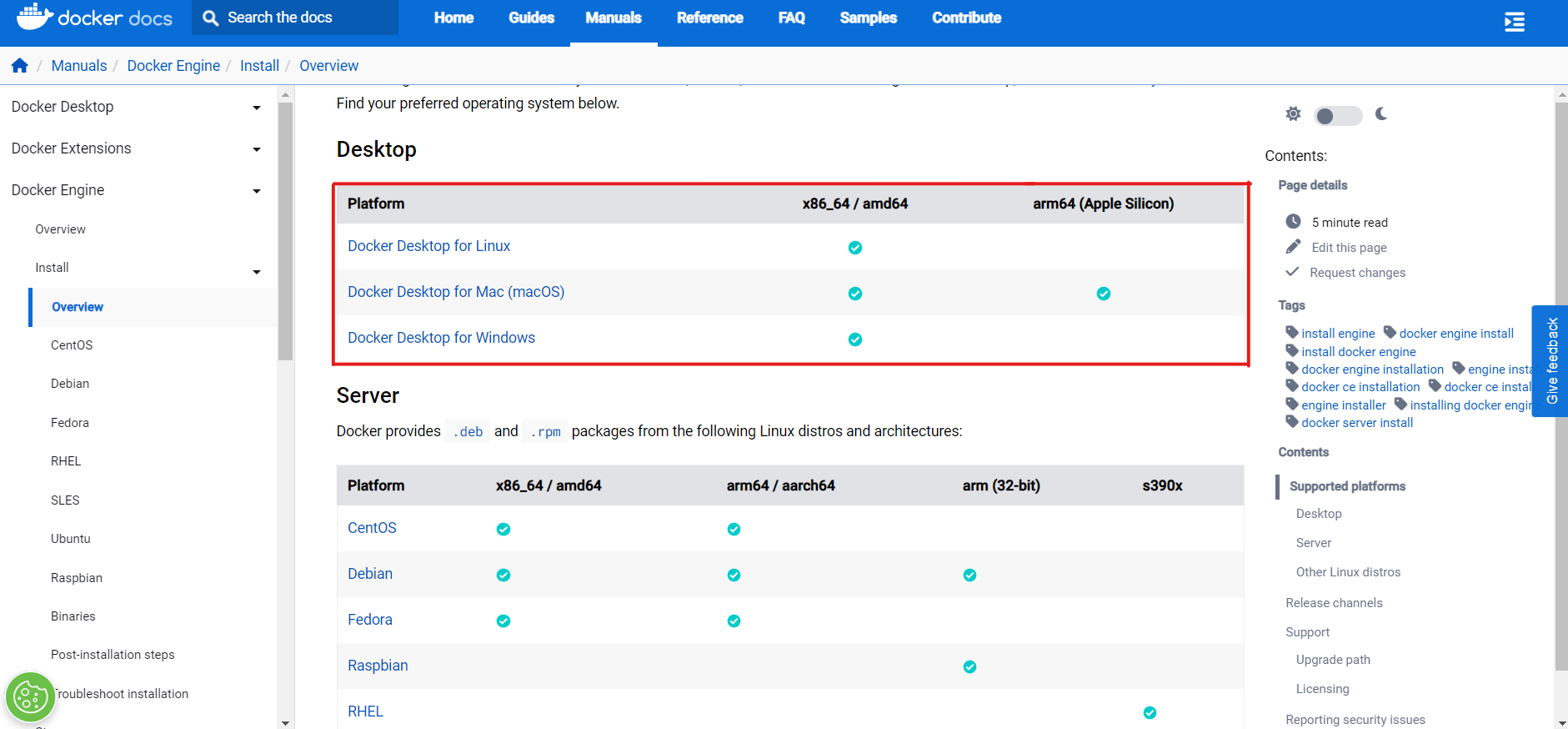
Follow the instructions on the appropriate page for the selected OS and install Docker.
Get Docker Image
- Pull image from Docker Hub.
docker pull xvykopal/monailabel-server:latest
Without Docker
General prerequisites
These prerequisites are necessary when you want to run the server without Docker.
Python
First of all, you need to install Python with the verion >=3.9 and setup the Python in your computer's settings.
Install Python version >= 3.9.
Add the path to Python to the environment variables. The path to Python added to the environment variables should look similar to the following:
C:\Users\{user}\AppData\Local\Programs\Python\{python}, where{user}is replaced with the user identifier within the OS and {python} represents the directory name with the installed Python for a specific version.2.1. On Windows, open the Start Menu and type, e.g
environment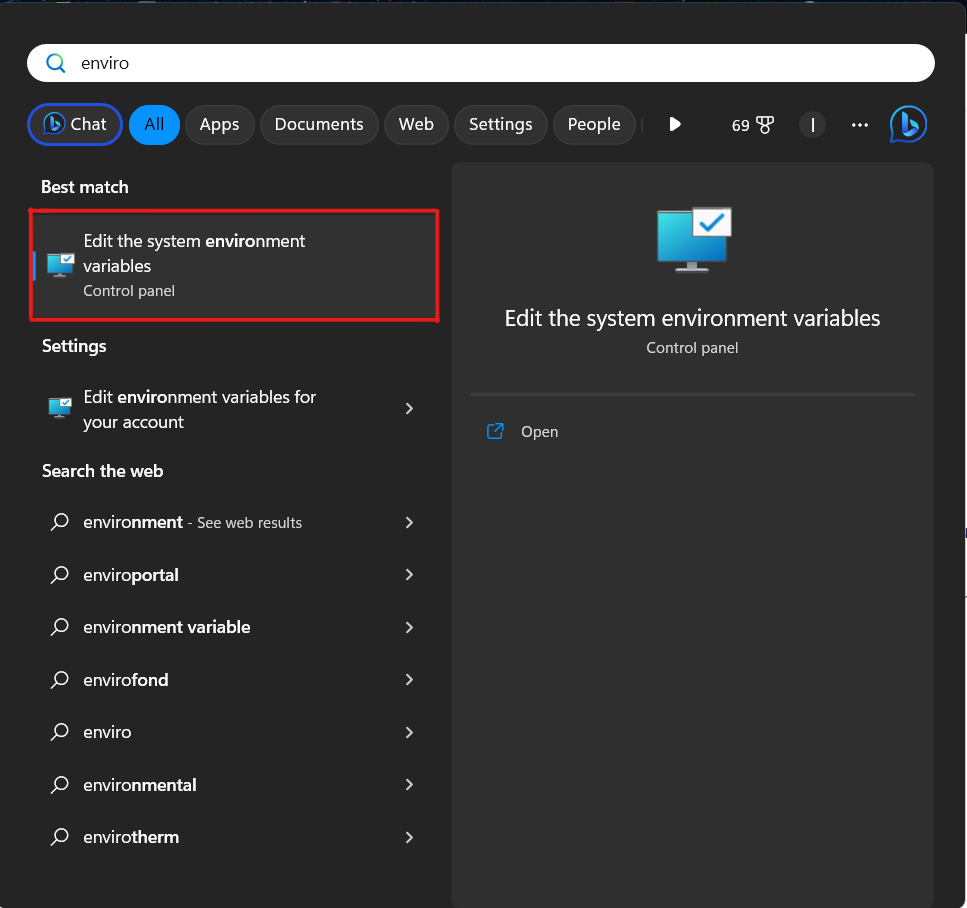
2.2 Select
Edit the system envirnoment variables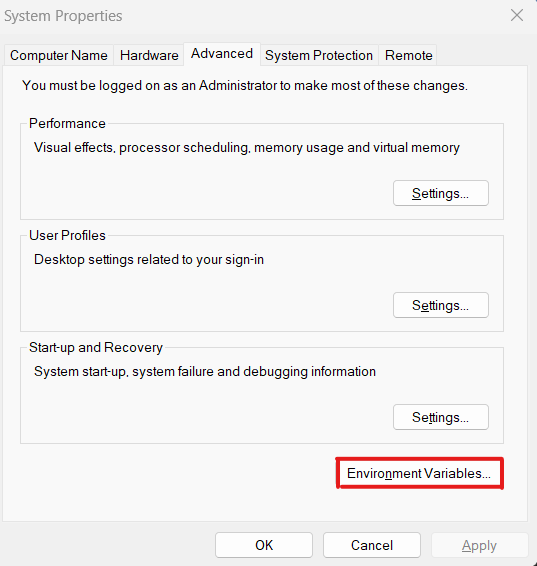
2.3. Edit
Pathvariable mainly for the user, but sometimes it is necessary to editPathvariable also in system variables.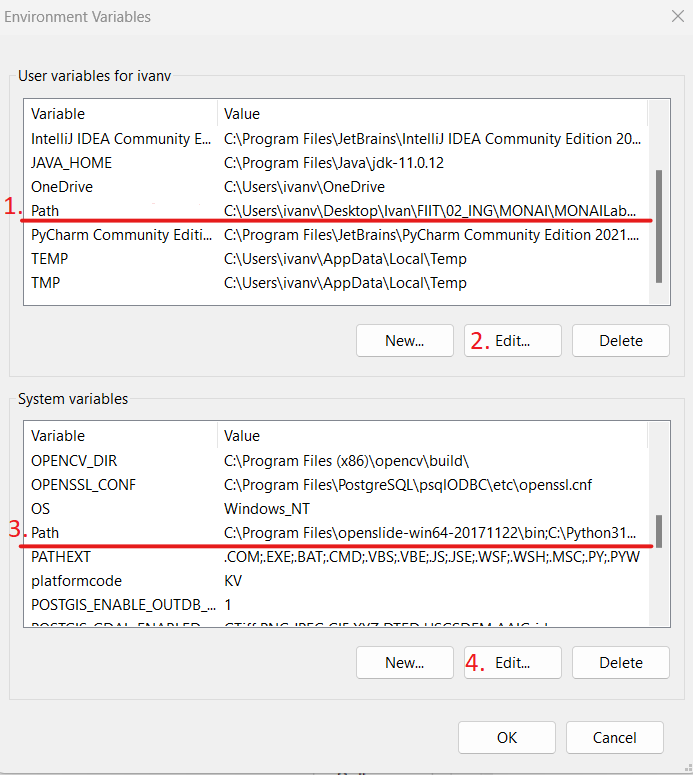
Python should be now available. You can check it in console by running the following command:
python --version, if there is no error it means that Python was successfully installed.
Source code
The next step is to download source code from the GitHub repository.
Download the development branch from the repository.
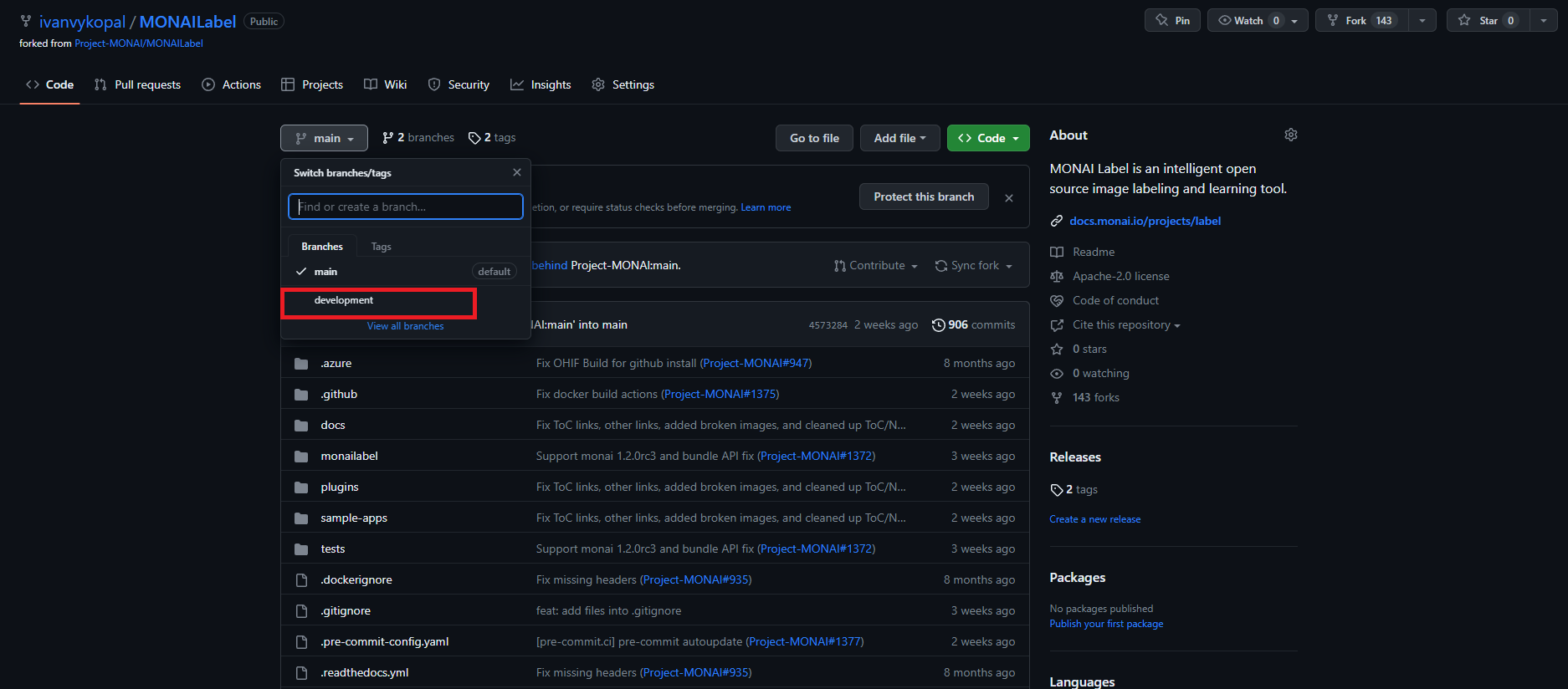
- The first method via
Code>Download ZIP
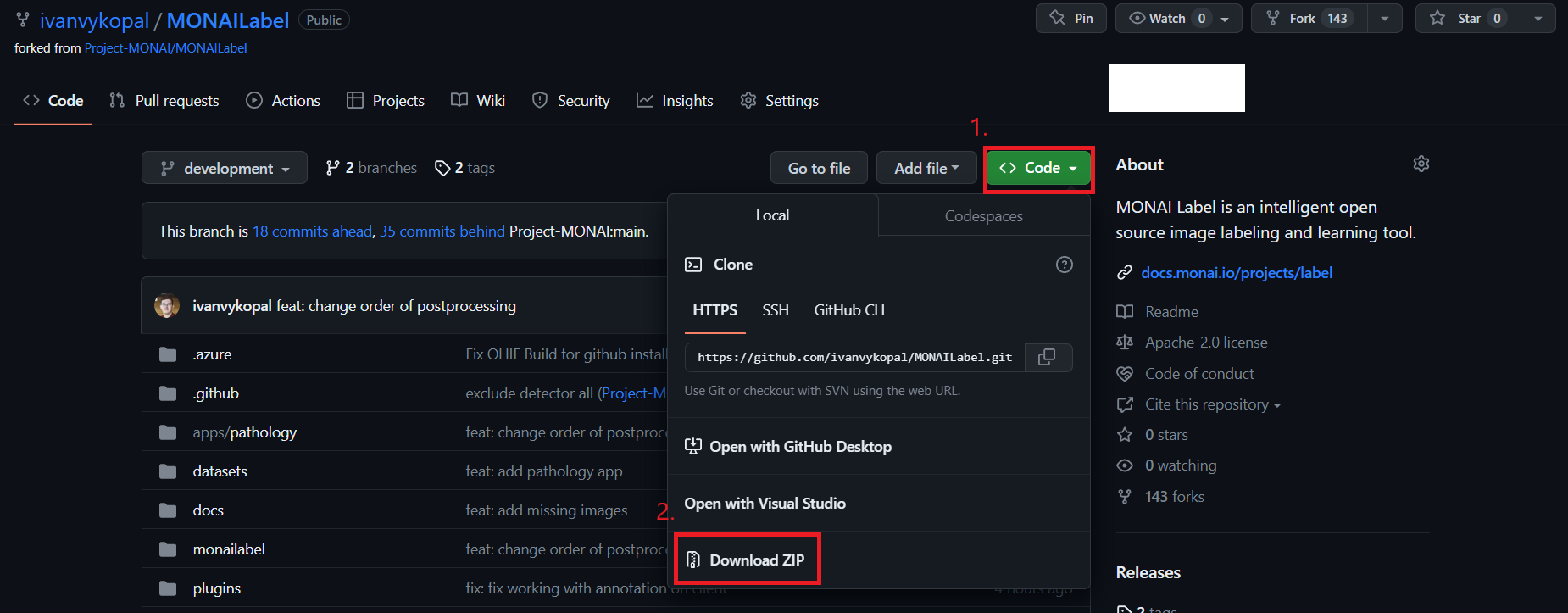
The second method via:
git clone https://github.com/ivanvykopal/MONAILabel.git
git fetch --all
git checkout development
- The first method via
After we downloaded the source code, we can create a new python environment:
Install the packages from
requirements.txtvenv.venv/monailabel/Scripts/activate
python -m pip install --upgrade pip
pip install -r requirements.txtcondaconda activate monailabel
python -m pip install --upgrade pip
pip install -r requirements.txt
To run server, it is needed to download openslide binaries and extract them to any location. After extraction, add the path to the
bindirectory to the environment variables inPATH, as you done for Python.To add the path to the
monailabel\scriptsdirectory to thePATHenvironment variable, follow the instructions in this macOS example. If anydlllibraries are missing, you can download them from dll-files.com. Specifically, if you need the missingcudnn64_8.dll, you can download it using this link. After downloading thedllfiles, you need to copy them to the system disk. On Windows systems, the standard location isC:\Windows\System32.
Weights and Sample Project
To run the inference of the added models, we need to download weigths of the models. To download them, it is necessary to contact Ivan Vykopal or VGG Research group
Download the weights of the models and sample project provided by authors.
Add the downloaded weights for
pathology_structure_segmentation_nestedunettoapps/pathology/model/pathology_structure_segmentation_nestedunet/modelsand add the weights forpathology_structure_segmentation_deeplabv3plustoapps/pathology/model/pathology_structure_segmentation_deeplabv3plus/models.